Using nsidc-iceflow with icepyx to Generate an Elevation Timeseries¶
This notebook shows how to produce a harmonized elevation timeseries across all iceflow-supported datasets along with ICESat-2 data using icepyx.
If this is your first time using nsidc-iceflow, we recommend starting with the NSIDC Iceflow example Jupyter Notebook first.
Similarly, if you are new to icepyx, we suggest reviewing the icepyx documentation for more information about how to use icepyx.
Scenario: assessing ice surface elevation change near Sermeq Kujalleq (Jakobshavn Isbrae)¶
In this notebook, we will focus on a small area near Sermeq Kujalleq (Jakobshavn Isbrae).
This notebook will demonstrate how to:
Search for and download all iceflow-supported data for our area of interest and timeframe.
Search for and download ICESat-2 data using
icepyx.Average elevation data over our area of interest at a weekly resolution.
Plot the results as a timeseries.
Import required packages¶
We will import several packages needed to perform our analysis:
# Imports
from __future__ import annotations
import datetime as dt
from pathlib import Path
import dask.dataframe as dd
import icepyx as ipx
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import xarray as xr
from nsidc.iceflow import (
IceflowDataFrame,
download_iceflow_results,
find_iceflow_data,
make_iceflow_parquet,
)
Search for and download iceflow-supported data for our area of interest and timeframe.¶
We first need to define some constants, including our download path, ITRF, and filter parameters. These will be used throughout the rest of the notebook.
Note that the data supported by nsidc-iceflow and icepyx can be very large! This tutorial will download ~2GB worth of data to local disk.
# Constants
# All of our data will be downloaded to this location.
OUTPUT_DIR = Path("./downloaded-data/")
OUTPUT_DIR.mkdir(exist_ok=True)
# ICESat2 data products use ITRF2014 (e.g., see https://nsidc.org/data/atl06/versions/6):
# > WGS 84 ellipsoid, ITRF2014 reference frame
# NOTE/TODO: This is expected to change in the near future! ICESat2 release
# 7, scheduled for spring 2025, is expected to be referenced to ITRF2020.
ICESAT2_ITRF = "ITRF2014"
# This bounding box covers an area near Sermeq Kujalleq (Jakobshavn Isbrae)
BBOX = (
-49.149,
69.186,
-48.949,
69.238,
)
# Range of dates we want to evaluate
DATE_RANGE = (dt.date(2007, 1, 1), dt.date(2024, 10, 28))
Next we will use the find_iceflow_data function from the nsidc-iceflow API to find data matching our area of interest.
By default, find_iceflow_data will include all nsidc-iceflow supported datasets, unless one or more are specified as a filter with the datasets kwarg. There may be warnings raised about there not being search results for specific datasets supported by nsidc-iceflow.
search_results = find_iceflow_data(
bounding_box=tuple(BBOX),
temporal=DATE_RANGE,
)
len(search_results)
2025-06-16 10:23:28.163 | WARNING | nsidc.iceflow.data.fetch:_find_iceflow_data_for_dataset:43 - Found no results for dataset.short_name='ILVIS2' dataset.version='1' with search_kwargs={'bounding_box': (-49.149, 69.186, -48.949, 69.238), 'temporal': (datetime.date(2007, 1, 1), datetime.date(2024, 10, 28))}
6
Now download the matching files to disk using download_iceflow_results.
Note: This next step may take a while, and will download data to your local disk.
downloaded_files = download_iceflow_results(search_results, output_dir=OUTPUT_DIR)
2025-06-16 10:23:34.292 | INFO | nsidc.iceflow.data.fetch:_download_iceflow_search_result:62 - Downloading 4 granules to downloaded-data/ILATM1B_1.
2025-06-16 10:23:53.408 | INFO | nsidc.iceflow.data.fetch:_download_iceflow_search_result:62 - Downloading 14 granules to downloaded-data/ILATM1B_2.
2025-06-16 10:24:08.263 | INFO | nsidc.iceflow.data.fetch:_download_iceflow_search_result:62 - Downloading 13 granules to downloaded-data/BLATM1B_1.
2025-06-16 10:24:57.489 | INFO | nsidc.iceflow.data.fetch:_download_iceflow_search_result:62 - Downloading 2 granules to downloaded-data/ILVIS2_2.
2025-06-16 10:25:13.304 | INFO | nsidc.iceflow.data.fetch:_download_iceflow_search_result:62 - Downloading 8 granules to downloaded-data/GLAH06_034.
Finally, we will create a parquet dataset using make_iceflow_parquet.Writing data to a parquet dataset allows dask (which we will use later!) to read the chunks of data it needs to do calculations (e.g., mean) without needing to read all of the data into memory at once. This is important because nsidc-iceflow can find many millions of data points for even small areas of interest!
This function transforms all data found in the data_dir to the provided target_itrf, further facilitating analysis across many datasets.
Note: This next step may take a while
parquet_path = make_iceflow_parquet(
data_dir=OUTPUT_DIR,
target_itrf=ICESAT2_ITRF,
overwrite=True,
)
Now we read the data stored in the parquet dataset using dask:
iceflow_df = dd.read_parquet(parquet_path)
# Ensure that our index is set as a datetime object
iceflow_df = iceflow_df.reset_index()
iceflow_df["utc_datetime"] = dd.to_datetime(iceflow_df["utc_datetime"])
iceflow_df = iceflow_df.set_index("utc_datetime")
iceflow_df.head()
| latitude | longitude | elevation | dataset | |
|---|---|---|---|---|
| utc_datetime | ||||
| 2007-03-12 06:39:58.970415 | 50.105011 | -41.965750 | 40.621659 | GLAH06v034 |
| 2007-03-12 06:39:58.995415 | 50.106557 | -41.966118 | 39.633659 | GLAH06v034 |
| 2007-03-12 06:39:59.220415 | 50.120483 | -41.969395 | 40.284658 | GLAH06v034 |
| 2007-03-12 06:39:59.320415 | 50.126654 | -41.970829 | 40.168658 | GLAH06v034 |
| 2007-03-12 06:40:00.520415 | 50.200840 | -41.988417 | 39.976656 | GLAH06v034 |
Search for and download ICESat-2 data using icepyx¶
Next, we will use icepyx to find ATL06 data for the same area of interest and timeframe.
To learn more about icepyx.Query, which is used below, see the documentation.
# We will compare the ILATM1B data to ATL06 data from October 2018.
result = ipx.Query(
"ATL06",
list(BBOX),
DATE_RANGE,
)
result
<icepyx.core.query.Query at 0x7b8227f62f00>
Now we place an order and download the results. Note that this may take a while to download data to local disk.
Note: These next steps may take a while
result.order_granules(subset=True)
Harmony job ID: 1d2cafe2-2691-495c-b00d-692624a72ddb
Initial status of your harmony order request: running
| Job ID | Type | Status | Details |
|---|---|---|---|
| 1d2cafe2-2691-495c-b00d-692624a72ddb | subset | running | View Details |
result.download_granules("downloaded-data/ATL06/")
Your harmony job status is still running. Please continue waiting... this may take a few moments.
................................Downloading results for harmony job 1d2cafe2-2691-495c-b00d-692624a72ddb
downloaded-data/ATL06/102107183_ATL06_20181111054901_06660105_006_02_subsetted.h5
downloaded-data/ATL06/102107184_ATL06_20181217152316_12220103_006_02_subsetted.h5
downloaded-data/ATL06/102107185_ATL06_20190112025232_02240205_006_02_subsetted.h5
downloaded-data/ATL06/102107187_ATL06_20190318110313_12220203_006_02_subsetted.h5
downloaded-data/ATL06/102107186_ATL06_20190115135914_02770203_006_02_subsetted.h5
downloaded-data/ATL06/102107190_ATL06_20190511210825_06660305_006_02_subsetted.h5
downloaded-data/ATL06/102107191_ATL06_20190617064249_12220303_006_02_subsetted.h5
downloaded-data/ATL06/102107193_ATL06_20190810164806_06660405_006_02_subsetted.h5
downloaded-data/ATL06/102107194_ATL06_20190916022239_12220403_006_02_subsetted.h5
downloaded-data/ATL06/102107195_ATL06_20191011135158_02240505_006_02_subsetted.h5
downloaded-data/ATL06/102107199_ATL06_20200110093142_02240605_006_01_subsetted.h5
downloaded-data/ATL06/102107197_ATL06_20191109122800_06660505_006_01_subsetted.h5
downloaded-data/ATL06/102107198_ATL06_20191215220228_12220503_006_01_subsetted.h5
downloaded-data/ATL06/102107201_ATL06_20200208080743_06660605_006_01_subsetted.h5
downloaded-data/ATL06/102107202_ATL06_20200315174217_12220603_006_01_subsetted.h5
downloaded-data/ATL06/102107203_ATL06_20200410051129_02240705_006_02_subsetted.h5
downloaded-data/ATL06/102107206_ATL06_20200614132201_12220703_006_01_subsetted.h5
downloaded-data/ATL06/102107205_ATL06_20200509034734_06660705_006_01_subsetted.h5
downloaded-data/ATL06/102107209_ATL06_20200807232719_06660805_006_01_subsetted.h5
downloaded-data/ATL06/102107211_ATL06_20201008203102_02240905_006_01_subsetted.h5
downloaded-data/ATL06/102107210_ATL06_20200913090149_12220803_006_02_subsetted.h5
downloaded-data/ATL06/102107214_ATL06_20210107161057_02241005_006_01_subsetted.h5
downloaded-data/ATL06/102107213_ATL06_20201213044139_12220903_006_01_subsetted.h5
downloaded-data/ATL06/102107212_ATL06_20201106190706_06660905_006_01_subsetted.h5
downloaded-data/ATL06/102107216_ATL06_20210205144704_06661005_006_01_subsetted.h5
downloaded-data/ATL06/102107217_ATL06_20210314002135_12221003_006_01_subsetted.h5
downloaded-data/ATL06/102107218_ATL06_20210408115050_02241105_006_02_subsetted.h5
downloaded-data/ATL06/102107222_ATL06_20210708073039_02241205_006_01_subsetted.h5
downloaded-data/ATL06/102107220_ATL06_20210507102653_06661105_006_01_subsetted.h5
downloaded-data/ATL06/102107224_ATL06_20210806060645_06661205_006_01_subsetted.h5
downloaded-data/ATL06/102107221_ATL06_20210612200124_12221103_006_01_subsetted.h5
downloaded-data/ATL06/102107225_ATL06_20211007031039_02241305_006_01_subsetted.h5
downloaded-data/ATL06/102107227_ATL06_20211105014647_06661305_006_01_subsetted.h5
downloaded-data/ATL06/102107229_ATL06_20220105225028_02241405_006_01_subsetted.h5
downloaded-data/ATL06/102107228_ATL06_20211211112116_12221303_006_01_subsetted.h5
downloaded-data/ATL06/102107234_ATL06_20220611024058_12221503_006_01_subsetted.h5
downloaded-data/ATL06/102107232_ATL06_20220312070106_12221403_006_01_subsetted.h5
downloaded-data/ATL06/102107231_ATL06_20220203212633_06661405_006_01_subsetted.h5
downloaded-data/ATL06/102107233_ATL06_20220505170624_06661505_006_01_subsetted.h5
downloaded-data/ATL06/102107237_ATL06_20220804124627_06661605_006_01_subsetted.h5
downloaded-data/ATL06/102107238_ATL06_20220909222056_12221603_006_01_subsetted.h5
downloaded-data/ATL06/102107241_ATL06_20221103082608_06661705_006_01_subsetted.h5
downloaded-data/ATL06/102107239_ATL06_20221005095006_02241705_006_01_subsetted.h5
downloaded-data/ATL06/102107235_ATL06_20220706141022_02241605_006_02_subsetted.h5
downloaded-data/ATL06/102107242_ATL06_20221209180033_12221703_006_02_subsetted.h5
downloaded-data/ATL06/102107243_ATL06_20230104052948_02241805_006_02_subsetted.h5
downloaded-data/ATL06/102107248_ATL06_20230503234538_06661905_006_03_subsetted.h5
downloaded-data/ATL06/102107245_ATL06_20230202040603_06661805_006_02_subsetted.h5
downloaded-data/ATL06/102107246_ATL06_20230405010947_02241905_006_02_subsetted.h5
downloaded-data/ATL06/102107253_ATL06_20230908045912_12222003_006_02_subsetted.h5
downloaded-data/ATL06/102107249_ATL06_20230609091949_12221903_006_02_subsetted.h5
downloaded-data/ATL06/102107252_ATL06_20230802192451_06662005_006_02_subsetted.h5
downloaded-data/ATL06/102107255_ATL06_20231101150428_06662105_006_01_subsetted.h5
downloaded-data/ATL06/102107258_ATL06_20240102120809_02242205_006_01_subsetted.h5
downloaded-data/ATL06/102107261_ATL06_20240402074729_02242305_006_01_subsetted.h5
downloaded-data/ATL06/102107263_ATL06_20240501062333_06662305_006_01_subsetted.h5
downloaded-data/ATL06/102107260_ATL06_20240131104358_06662205_006_01_subsetted.h5
downloaded-data/ATL06/102107264_ATL06_20240702032717_02242405_006_01_subsetted.h5
downloaded-data/ATL06/102107257_ATL06_20231208003854_12222103_006_03_subsetted.h5
downloaded-data/ATL06/102107268_ATL06_20240930230647_02242505_006_02_subsetted.h5
downloaded-data/ATL06/102107266_ATL06_20240731020304_06662405_006_01_subsetted.h5
downloaded-data/ATL06/102107267_ATL06_20240905113742_12222403_006_01_subsetted.h5
[PosixPath('downloaded-data/ATL06/102107183_ATL06_20181111054901_06660105_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107184_ATL06_20181217152316_12220103_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107185_ATL06_20190112025232_02240205_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107187_ATL06_20190318110313_12220203_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107186_ATL06_20190115135914_02770203_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107190_ATL06_20190511210825_06660305_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107191_ATL06_20190617064249_12220303_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107193_ATL06_20190810164806_06660405_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107194_ATL06_20190916022239_12220403_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107195_ATL06_20191011135158_02240505_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107199_ATL06_20200110093142_02240605_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107197_ATL06_20191109122800_06660505_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107198_ATL06_20191215220228_12220503_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107201_ATL06_20200208080743_06660605_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107202_ATL06_20200315174217_12220603_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107203_ATL06_20200410051129_02240705_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107206_ATL06_20200614132201_12220703_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107205_ATL06_20200509034734_06660705_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107209_ATL06_20200807232719_06660805_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107211_ATL06_20201008203102_02240905_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107210_ATL06_20200913090149_12220803_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107214_ATL06_20210107161057_02241005_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107213_ATL06_20201213044139_12220903_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107212_ATL06_20201106190706_06660905_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107216_ATL06_20210205144704_06661005_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107217_ATL06_20210314002135_12221003_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107218_ATL06_20210408115050_02241105_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107222_ATL06_20210708073039_02241205_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107220_ATL06_20210507102653_06661105_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107224_ATL06_20210806060645_06661205_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107221_ATL06_20210612200124_12221103_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107225_ATL06_20211007031039_02241305_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107227_ATL06_20211105014647_06661305_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107229_ATL06_20220105225028_02241405_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107228_ATL06_20211211112116_12221303_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107234_ATL06_20220611024058_12221503_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107232_ATL06_20220312070106_12221403_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107231_ATL06_20220203212633_06661405_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107233_ATL06_20220505170624_06661505_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107237_ATL06_20220804124627_06661605_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107238_ATL06_20220909222056_12221603_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107241_ATL06_20221103082608_06661705_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107239_ATL06_20221005095006_02241705_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107235_ATL06_20220706141022_02241605_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107242_ATL06_20221209180033_12221703_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107243_ATL06_20230104052948_02241805_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107248_ATL06_20230503234538_06661905_006_03_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107245_ATL06_20230202040603_06661805_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107246_ATL06_20230405010947_02241905_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107253_ATL06_20230908045912_12222003_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107249_ATL06_20230609091949_12221903_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107252_ATL06_20230802192451_06662005_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107255_ATL06_20231101150428_06662105_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107258_ATL06_20240102120809_02242205_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107261_ATL06_20240402074729_02242305_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107263_ATL06_20240501062333_06662305_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107260_ATL06_20240131104358_06662205_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107264_ATL06_20240702032717_02242405_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107257_ATL06_20231208003854_12222103_006_03_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107268_ATL06_20240930230647_02242505_006_02_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107266_ATL06_20240731020304_06662405_006_01_subsetted.h5'),
PosixPath('downloaded-data/ATL06/102107267_ATL06_20240905113742_12222403_006_01_subsetted.h5')]
Read ICESat-2 data into xarray and pandas¶
Next, we will use icepyx.Read to read the data into an xarray Dataset.
Note that the code below wraps the reading of each file in a try/except block because of a known issue with subsetting. See https://github.com/icesat2py/icepyx/issues/576 for more information.
datasets = []
for file in Path("downloaded-data/ATL06/").glob("*.h5"):
try:
reader = ipx.Read(str(file))
reader.variables.append(var_list=["h_li", "latitude", "longitude"])
ds = reader.load()
datasets.append(ds)
except Exception:
print(f"{file=} contains an error and will not be read")
continue
len(datasets)
file=PosixPath('downloaded-data/ATL06/102107201_ATL06_20200208080743_06660605_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107263_ATL06_20240501062333_06662305_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107238_ATL06_20220909222056_12221603_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107186_ATL06_20190115135914_02770203_006_02_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107264_ATL06_20240702032717_02242405_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107214_ATL06_20210107161057_02241005_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107248_ATL06_20230503234538_06661905_006_03_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107224_ATL06_20210806060645_06661205_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107234_ATL06_20220611024058_12221503_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107213_ATL06_20201213044139_12220903_006_01_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107268_ATL06_20240930230647_02242505_006_02_subsetted.h5') contains an error and will not be read
file=PosixPath('downloaded-data/ATL06/102107253_ATL06_20230908045912_12222003_006_02_subsetted.h5') contains an error and will not be read
50
ds = xr.concat(datasets, dim="gran_idx")
ds.head()
<xarray.Dataset> Size: 5kB
Dimensions: (gran_idx: 5, spot: 5, photon_idx: 5)
Coordinates:
* photon_idx (photon_idx) int64 40B 0 1 2 3 4
* spot (spot) uint8 5B 1 2 3 4 5
* gran_idx (gran_idx) uint64 40B 22412 66614 122213 22401 66610
source_file (gran_idx) <U81 2kB 'downloaded-data/ATL06/102107225...
delta_time (gran_idx, photon_idx) datetime64[ns] 200B 2021-10-0...
Data variables:
sc_orient (gran_idx) int8 5B 0 0 0 0 1
cycle_number (gran_idx) int8 5B 13 15 14 2 11
rgt (gran_idx) int16 10B 224 666 1222 224 666
atlas_sdp_gps_epoch (gran_idx) datetime64[ns] 40B 2018-01-01T00:00:18 .....
data_start_utc (gran_idx) datetime64[ns] 40B 2021-10-07T03:10:39.42...
data_end_utc (gran_idx) datetime64[ns] 40B 2021-10-07T03:15:15.96...
h_li (spot, gran_idx, photon_idx) float32 500B 793.6 ... nan
latitude (spot, gran_idx, photon_idx) float64 1kB 69.24 ... nan
longitude (spot, gran_idx, photon_idx) float64 1kB -49.15 ... nan
gt (gran_idx, spot) object 200B 'gt1l' nan ... 'gt2l' nan
Attributes:
data_product: ATL06
Description: The land_ice_height group contains the primary set of deri...
data_rate: Data within this group are sparse. Data values are provid...icepyx reads ICESat-2 data as an xarray dataset. xarray is a powerful tool and the data is ready to use in this format, but to simplify things for this notebook and make the data more compatible with nsidc-iceflow, the next step will convert the data into an nsidc-iceflow-compatible pandas DataFrame.
The first step is to rename the variables to be consistent with how nsidc-iceflow identifies the elevation and time fields:
ds = ds.rename({"h_li": "elevation", "delta_time": "utc_datetime"})
The following code block converts the xarray dataset into an iceflow dataframe:
spot_dfs = []
for spot in ds.spot.data.flatten():
df = pd.DataFrame.from_dict(
{
"elevation": ds.sel(spot=spot).elevation.data.flatten(),
"latitude": ds.sel(spot=spot).latitude.data.flatten(),
"longitude": ds.sel(spot=spot).longitude.data.flatten(),
"utc_datetime": ds.sel(spot=spot).utc_datetime.data.flatten(),
"spot": [spot] * len(ds.sel(spot=spot).elevation.data.flatten()),
"ITRF": ICESAT2_ITRF,
},
)
spot_dfs.append(df)
df = pd.concat(spot_dfs, sort=True)
# Drop rows where lat, lon, or elev are missing.
df = df.dropna(subset=["latitude", "longitude", "elevation"], how="any")
df = df.set_index("utc_datetime")
# Cast the df as an IceflowDataFrame.
# The `atl06_df` can now be used with e.g., `nsidc-iceflow`'s ITRF conversion function
# to perform plate motion model adjustments if desired.
atl06_df = IceflowDataFrame(df)
atl06_df.head()
| ITRF | elevation | latitude | longitude | spot | |
|---|---|---|---|---|---|
| utc_datetime | |||||
| 2021-10-07 03:13:30.711221536 | ITRF2014 | 793.648804 | 69.237833 | -49.146681 | 1 |
| 2021-10-07 03:13:30.714035872 | ITRF2014 | 793.453491 | 69.237655 | -49.146743 | 1 |
| 2021-10-07 03:13:30.716850784 | ITRF2014 | 793.404785 | 69.237477 | -49.146805 | 1 |
| 2021-10-07 03:13:30.719666512 | ITRF2014 | 793.342651 | 69.237299 | -49.146866 | 1 |
| 2021-10-07 03:13:30.722483664 | ITRF2014 | 793.249939 | 69.237121 | -49.146928 | 1 |
The ATL06 data contains some negative elevation values, as we see printed below. We will filter these out, as we expect positive elevations.
print(atl06_df.elevation.min())
# Filter out negative values. We expect positive elevations.
atl06_df = atl06_df[atl06_df.elevation > 0]
-1320.41162109375
Average elevation data over our area of interest at a weekly resolution¶
In this next step, we will resample the ATL06 and nsidc-iceflow data to a weekly resolution, taking the mean of elevation over our area of interest. This will provide us with one data point per week where data is available, giving us a general idea of how the elevation of our area of interest changes over time. First we resample ATL06 data to a weekly mean:
atl06_avg_df = atl06_df[["elevation"]].resample("W").mean()
atl06_avg_df = atl06_avg_df.dropna(how="any")
atl06_avg_df.head()
| elevation | |
|---|---|
| utc_datetime | |
| 2018-11-11 | 876.653491 |
| 2018-12-23 | 887.003014 |
| 2019-01-13 | 836.233506 |
| 2019-03-24 | 865.448348 |
| 2019-05-12 | 877.559574 |
And now we resample the iceflow data to a weekly mean:
iceflow_df_sampled = iceflow_df.repartition(freq="1W")
iceflow_df_sampled = iceflow_df_sampled.dropna(how="any")
iceflow_df_sampled = iceflow_df_sampled[iceflow_df_sampled.elevation > 0]
iceflow_avg = iceflow_df_sampled.resample("W").agg(
{
"elevation": "mean",
"dataset": lambda x: ", ".join(x.astype("str").unique()),
}
)
iceflow_avg = iceflow_avg.replace([np.inf, -np.inf], np.nan)
iceflow_avg = iceflow_avg.dropna(how="any")
iceflow_avg = iceflow_avg.compute()
iceflow_avg.head()
| elevation | dataset | |
|---|---|---|
| utc_datetime | ||
| 2007-03-18 | 657.922769 | GLAH06v034 |
| 2007-10-07 | 438.062308 | GLAH06v034 |
| 2008-02-17 | 181.279161 | GLAH06v034 |
| 2008-06-29 | 888.758989 | BLATM1Bv1 |
| 2008-07-06 | 879.285982 | BLATM1Bv1 |
Plot the results as a timeseries¶
Now we will use matplotlib to plot the results as a timeseries:
%matplotlib inline
# We will use a unique marker for each dataset
iceflow_marker_map = {
"GLAH06v034": "s",
"BLATM1Bv1": "D",
"ILATM1Bv1": "x",
"ILATM1Bv2": "o",
"ILVIS2v1": "v",
"ILVIS2v2": "^",
}
fig = plt.figure(figsize=(10, 8))
for dataset, marker in iceflow_marker_map.items():
subset = iceflow_avg[iceflow_avg.dataset == dataset]
if subset.elevation.any():
plt.scatter(
subset.index,
subset.elevation,
marker=marker,
label=dataset,
linestyle="",
color="black",
)
plt.scatter(
atl06_avg_df.index,
atl06_avg_df.elevation,
color="black",
marker="*",
label="ATL06v6",
linestyle="",
)
plt.xlabel("Date")
plt.ylabel("Elevation")
plt.legend(title="Dataset")
<matplotlib.legend.Legend at 0x7b822655c500>
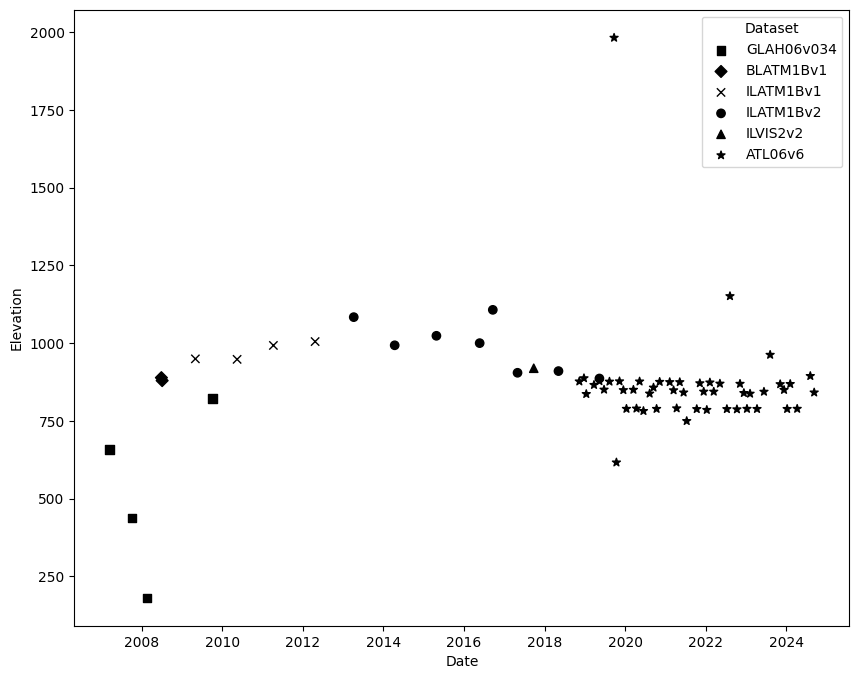
Conclusions¶
In this notebook, we found and analyzed laser altimetry data from a variety of datasets using nsidc-iceflow and icepyx.
In the timeseries plot above, we can see how the surface elevation of a small area near Sermeq Kujalleq (Jakobshavn Isbrae) changes over time.
Note that this analysis was relatively simple. Although the data and plot above give us an idea of surface elevation changes, it should be noted that there are still a number of things a researcher should consider when doing an analysis across many datasets over time. To further this analysis, we may want to consider doing one or more of the following:
Outlier detection and filtering
Cross-calibration of data between sensors/datasets
Plate motion model coordinate propagation (see the
nsidc-iceflowexample Jupyter Notebook for an example of how to do this)Account for spatial variability within our region of interest